08-Aug-2024
Nanobodies (Nbs) are small and stable fragments of heavy chain antibodies. Nbs are just single-domain fragments but have the same full antigen-binding capacity of the full antibody. Naturally occurring in camelids, the size and flexibility of Nbs makes them a critical tool in stabilising and “freezing” dynamic proteins into single conformations, to help characterise their structure.
Protein-protein interactions (PPIs) are a key example of a dynamic protein scenario, as changing conformations to allow for binding sees them take several forms. The research paper from Fischer et al (2024) describes the work conducted at VIB-VUB (Nanobodies4Instruct, part of Instruct-BE), utilising Nbs to stabilise the binding of SOS1 and KRAS in their signalling complex.
KRAS is a small GTPase that cycles between the GTP-loaded “on” state and the GDP-loaded “off” state. This is crucial for normal cell proliferation and survival and represent the most frequently mutated oncoprotein family in human cancers, with a reputation for being undruggable. One of the major regulators of this process is the Son of Sevenless (SOS) protein, a guanine nucleotide exchange factor that acts as a key activator for KRAS function.
The team at VIB-VUB identified four Nbs that either disrupt or stabilise the exchange of nucleotides in the SOS1•RAS signalling complex.
The first step was to Cross-link PPIs and Immunize llamas (ChILL). The SOS1•RAS complex was cross linked with glutaraldehyde to freeze the interacting proteins in a conformation very similar to the native PPI. Llamas were then immunised with this antigenic complex to develop the necessary Nbs. Following this, display and co-selection (DISCO) was applied, in which the immune libraries extracted from the ChILL process were displayed on yeast, with different coloured stains for Nbs that bound to the full SOS1•RAS complex, and those that bind individually. Two competitive binders that inhibit the SOS1•RAS association (Nb77 & Nb84) and two Nbs (Nb14 & Nb22) that bind the binary complex were fully characterised.
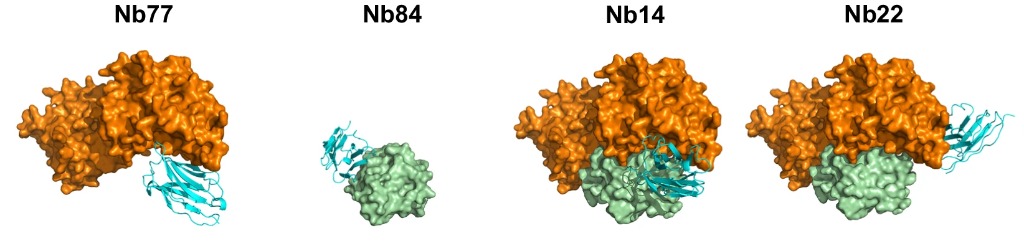
Fig 1. Crystal structures of the Nbs (cyan) in complex with SOS1 (orange) and/or RAS (green).
Nb77 binds to SOS1, blocking the binding site for RAS and preventing nucleotide exchange. Conversely, Nb84 binds to RAS itself, competing for the SOS1 binding site and again preventing nucleotide exchange. Nb14 and Nb22, however, bind to the complex itself (and also to SOS1, as confirmed by BLI) so they stabilise the PPI and promote nucleotide exchange. X-ray crystallography performed at Diamond Light Source (part of Instruct-UK) confirmed the binding site for these Nbs: Nb14 also connected to the RAS binding site of SOS1, but in a configuration that did not block the PPI formation, accelerating nucleotide exchange a stunning 27-fold. Nb22 binds SOS1 in a completely opposite epitope to the RAS catalytic site, increasing nucleotide exchange more than 2-fold. Of these, Nb14 was found to operate in a way more similar to natural allosteric modulators, compared to Nb22. Nb14 is found to change the conformation of the SOS1 binding site to bind RAS•GDP (which then will release the GDP, bind GTP, dissociate SOS1 and complete the reaction), speeding up the nucleotide exchange process two-fold.
Finally, the study utilised nanobodies to alter the signal intensity of SOS1•RAS•aminopiperidine indole (a complex previously found to accelerate nucleotide exchange) in NMR experimentation. They found that the addition of Nb22 significantly increased signal strength for NMR, making it a potentially vital addition for fragment-screening studies. This is of note for the Fragment-Screen project coordinated by Instruct, which is developing new tools and techniques for fragment-based drug discovery.
Overall, the study outlines the value of nanobody discovery, and how it can be used across several domains of drug discovery, and offers a novel approach to stabilising and characterising proteins as they interact with other proteins.