03-Oct-2024
Frizzled (FZD) receptors are transmembrane G-protein coupled receptors (GPCRs) activated by the binding of a ligand, Wnt, to an extracellular Cysteine Rich Domain (CRD). Signalling downstream of FZD receptors is fundamental to many different biological processes, and dysregulated signalling may play a role in Parkinson’s disease and Melanoma. FZD receptors signal through either canonical (β-catenin dependent) or non-canonical (primarily β-catenin independent) routes. Non-canonical receptors (e.g FZD3) are activated when a bivalent Wnt ligand simultaneously interacts with FZD and another receptor (most often a receptor tyrosine kinase). Understanding how these FZD receptors function requires insight into their interactions with Wnt ligands.
Using Instruct funded access to the VIB-VUB center for structural biology at Instruct-BE, Hillier & Zhao, et al were able to generate a series of FZD3 specific nanobodies. Their binding sites on the receptor were mapped using Biolayer Interferometry (BLI), with a nanobody dubbed ‘nanobody 8’ (Nb8) competing for the Wnt binding site within the extracellular CRD domain. Crystallisation and subsequent study of the structure of the Nb8 bound FZD3 CRD using X-ray crystallography (Diamond Light Source, Instruct-UK) revealed a 1.8Å structure with the key binding site usually occupied by Wnt ligands occluded by Nb8 (Figure 1A). The authors then assessed whether Nb8 could be functionalised as a Wnt surrogate by fusing the C-terminus of Dikkopf-1 (DKK1C) to the N-terminus of Nb8. DKK1C binds to the Wnt coreceptor LRP2/3, leading to downstream β-catenin dependent signalling upon binding of Nb8 to FZD3. The ability of Nb8_DKK1C to elicit a signalling response was assessed in an in vivo nanoluciferase assay that demonstrated that the Nb8_DKK1C fusion functioned as a Wnt surrogate capable of initiating β-catenin signalling.
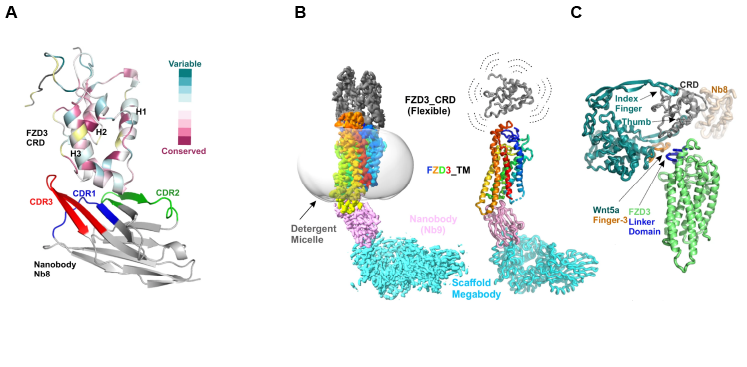
Figure 1: Structural characterisation of FZD3. A.) Structure of the FZD3 CRD bound to Nb8. FZD3 CRD helices 1-3 are labelled, as are the epitope binding regions (CDRs) of Nb8. Evolutionarily conserved and variable regions within FZD3 CRD are also highlighted. B.) Structure of the complete FZD3 receptor determined using cryo-EM and a Nb9 derived megabody. The CRD crystal structure resolved in A is fitted into the density in B. C.) Interface between FZD3 CRD and Nb8/Wnt5a. The Wnt5a Finger-3 contacts the FZD3 CRD as well as the linker domain.
In addition to Nb8, other Nanobodies generated and characterised in this study include Nb9, which binds to the intracellular portion of the FZD3 receptor. The fusion of Nb9 and bacterial α-galactosidase to produce a larger species termed a megabody enabled the structural characterisation of the complete FZD3 receptor by cryo-EM at 2.8Å global resolution (using microscopes at OPIC/eBIC, Instruct-UK) (Figure 1B). Whilst the FZD3 Transmembrane Domains (TMDs) were well resolved in this structure, revealing a small potentially druggable binding pocket also seen in FZD7, the flexible CRD was less well resolved. Fitting the CRD crystal structure previously generated in this study within this density, before superimposing the structure of the FZD8 CRD bound to Wnt5a allowed the modelling of Wnt5a on to this structure. This revealed that a hairpin loop within Wnt5a (dubbed ‘Finger 3’) made contact with both the CRD itself and the linker region connecting the CRD and TMD (Figure 1C). This interaction was confirmed by BLI, and AlphaFold (AF) modelling suggested that two amino acids in the hairpin loop that are poorly conserved between Wnt isoforms likely contribute to FZD selectivity.
Following FZD activation, Dishevelled (DVL) binds to an intracellular G-protein binding site on FZD3 and acts as a hub for several different signalling pathways. AF modelling reveals a predicted overlap between the binding sites of DVL and Nb9, verified subsequently in vitro via BLI, and in vivo, with Nb9 transfection blocking DVL2 recruitment to FZD3 at the plasma membrane. To test whether Nb9 binding to FZD3 also blocks the recruitment and coupling of G proteins at this binding site (as modelled in Figure 2A) Hillier & Zhao et al used an in vivo Bioluminescent Resonance Energy Transfer (BRET) assay. Using RLuc8 tagged GαS and GFP tagged Gγ, it was shown that Nb9 fusion to FZD3 leads to a disruption in basal FZD3 signalling by preventing the formation of a heterotrimeric G-protein complex (Figure 2B).
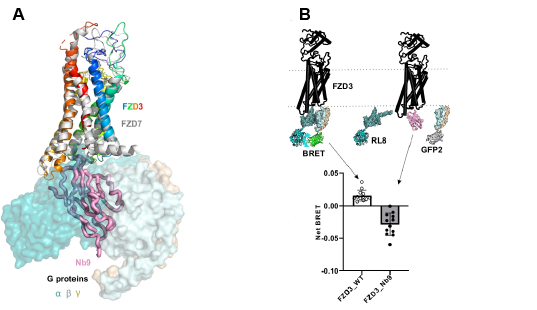
Figure 2: Nb9 as a FZD signalling blocker. A.) Schematic illustrating the Nb9 bound intracellular/TM portions of the FZD3 receptor overlayed with FZD7 bound to Gαβγ proteins. B.) Diagram describing the in vivo BRET-based approach used to illustrate that Nb9 can block intracellular β-catenin signalling. RL8 (RLuc8) and GFP2, the BRET biosensors, were attached to GαS and Gγ subunits. Transfection of Nb9 reduces FZD signalling to below basal levels.
By adopting an integrated structural biology approach involving multiple techniques available through Instruct-ERIC, Hillier & Zhao et al were able to generate high resolution structures of FZD3, as well as unveiling a key determinant to Wnt selectivity. In addition to this the nanobodies designed in this study to assist with structural characterisation show promise as modulators of FZD3 mediated signalling. On the support provided by Instruct-BE, principal investigator Professor E Yvonne Jones, said “Nanobodies played an essential role in our study; we were hugely impressed by the expertise and very grateful for the good-humored support and advice we received at every stage from colleagues at the VIB-VUB center for structural biology in Instruct-BE"