23-Oct-2024
In NF-κB signalling, phosphorylation and activation of the Inhibitor of κB Kinase (IKK) results in IKK-mediated phosphorylation of Inhibitor of κB (IκB), priming it for subsequent ubiquitination and degradation. This releases NF-κB dimers, transcription factors that are bound and cytosolically sequestered by IκB under basal conditions, so they can translocate to the nucleus to enact a variety of different transcriptional responses.
The canonical IKK complex consists of an IKKα/IKKβ heterodimer and two copies of the regulatory subunit NEMO. The non-canonical IKK complex consists solely of IKKα/IKKβ homodimers. Given that in the canonical IKK complex some specificity towards IκBα is conferred by NEMO, other mechanisms must exist to confer substrate specificity in the non-canonical complexes. In this study Li et al used an array of biochemical and integrated structural biology approaches to identify and characterise a Short Linear Motif (SLiM) facilitating the docking of IKKα/β dimers with several different partner proteins, including IκBα.
In vitro pulldown experiments firstly revealed that deletion of the IκBα C-terminus perturbs the IκBα IKKβ interaction. Further dissection of the C-terminus showed that a 17 amino acid motif containing 5 highly conserved residues was critical for binding. The interaction between peptides containing the YDDΦxΦ sequence (x denotes any amino acid, Φ denotes a hydrophobic residue) and IKKα/IKKβ homodimers purified from insect cells (using Instruct funded access to the protein production platform at Instruct-FR1, IGBMC Strasbourg) was assessed by Isothermal Calorimetry (ITC) at the ITC Advanced platform at IBMC, Strasbourg. Results showed significantly higher binding affinities to the motif for IKKα as compared to the IKKβ subunit. Furthermore, a C308L mutation at Φ4 resulted in increased binding affinity between IκBα and IKKα/IKKβ homodimers, which translated to an increase in IκBα phosphorylation and more rapid IκBα degradation in vivo. Y305 was also shown to be the sole phosphorylated residue between amino acid 73 and the IκBα C-terminus under basal conditions, with ITC revealing that Y305 phosphorylation prevents IκBa binding to IKKα, and thus acts as a switch regulating IKK binding.
To characterise this newly uncovered interaction Li et al used Instruct-funded access to the crystallisation facility at Instruct-FR1 and X-ray crystallography to produce a 4.16-6.18A structure of IKKβ (1-699) bound to a 17 amino acid IκBα peptide consisting of the YDDΦxΦ sequence and an N-terminal acidic patch. Whilst this resolution wasn’t sufficient to determine the peptide binding site, Crosslinking Mass Spectrometry (CLMS) showed that the motif peptide targeted the Scaffold Dimerisation Domains (SDDs) of IKK (Figure 1A). The in silico modelling tool HADDOCK (developed by the Bonvin Lab, Utrecht University, Instruct-NL), used in conjunction with distance constraints obtained by CLMS, suggested that the interaction occurred at the interface of the SDD domains in the IKK dimer (Figure 1B). This binding groove contains many polar/charged residues alongside several hydrophobic residues that form a pocket within which the hydrophobic residues within the YDDΦxΦ motif are inserted. These findings were validated in vitro and in vivo using pulldowns and the Gaussia princeps luciferase complementation assay (GPCA), by making mutations that disrupt IKK dimerisation or introduce charge reversal at key sites that attenuate IκBα binding.
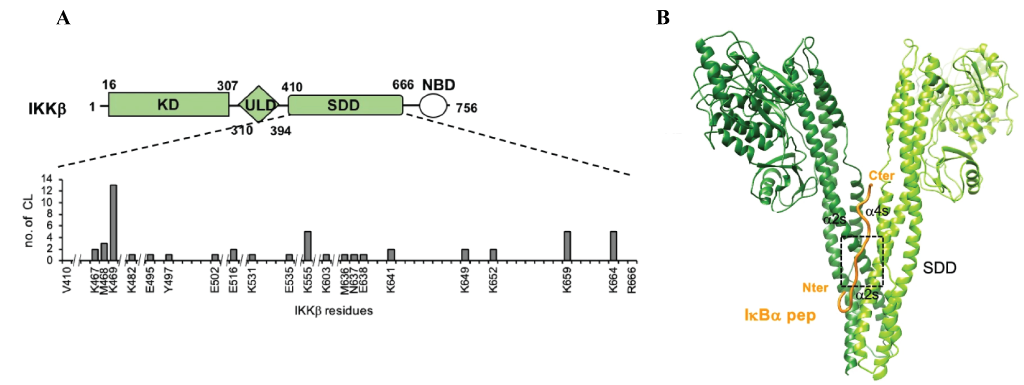
Figure 1: Interaction between IKKβ and IκBα peptide. A.) Schematic illustrating the crosslinks between IκBα with IKKβ are within the SDD domain, rather than the Kinase domain (KD) or Ubiquitin like domain (ULD). B.) Model of IKKβ bound to IκBα peptide based on the crystal structure obtained here for IKKβ, with IκBα peptide modelled using distance constraints from cross linking mass spectrometry.
Several other known interactors (IkBβ, p100, and IRF7) contain the YDDΦxΦ consensus motif. Using ITC and synthetic peptides based on the consensus motif in each interactor, Li et al showed that all the peptides interacted with IKKα, indicating a common mechanism of interaction between these proteins. In contrast IKKβ only interacted with IkB derived peptides (i.e. IkBα and IkBβ, not p100 and IRF7), indicating subtle variations in the binding specificities of the two catalytic subunits.
In summary Li et al reveal a novel SLiM recognised by IKK homo/heterodimers via a dimerisation dependent mechanism that is negatively regulated by SLiM phosphorylation. The authors also showed that synthetic peptides containing the C308L mutation block IκBα phosphorylation and degradation in cellulo, potentially highlighting a novel route to IKK inhibition in pathologies arising from NF-κB dysregulation. Instruct funded access to facilities at Instruct FR1 for insect cell expression and crystallisation trials provided the means for several of these experiments, whilst accessing the advanced ITC platform at IBMC Strasbourg enabled the authors to conduct important ITC experiments.
Principal investigator Katia Zanier said ‘It is always an enriching experience to interact with my colleagues at the INSTRUCT platforms in Strasbourg. Studying protein-protein interactions in signalling can be challenging due to the transient affinities and this is why we often need to combine several approaches. Hence, the access to the INSTRUCT infrastructure and to the know how of the platform scientists and engineers is vital for the success of this type of projects’.