16-Dec-2024
Flotillins are fundamental proteins in eukaryotic cells for membrane function, specifically for the formation of microdomains or lipid rafts, which are then central to cell signalling and trafficking. It is increasingly clear that flotillins, and in particular FloA, or flotillin, are also an integral part of the bacterial membrane with flotillin accumulating at functional membrane microdomains (FMMs), enriched in staphyloxanthin (STX). The team from the CNB-CSIC microbiology facility (Ukleja et al, 2024) sought to understand how flotillin affects bacterial pathogenicity, and the structure of its integration in the cell membrane by examining it in S. aureus, the bacteria behind MRSA.
The team first confirmed that flotillin, along with its functional partner membrane protein NfeD, are crucial for bacterial survival in stressful conditions. Mutation of both proteins (FloA and NfeD) and assessment of the number of colonies formed revealed a significant reduction in survivability for mutated strains compared to wild-type (WT). The structure of FloA was examined using AlphaFold 2, with predictions showing that FloA N terminus is adjacent to a cytoplasmic prohibitin domain (PHB). The PHB contains a subdomain of six parallel α-helices, as well as a coiled-coil domain (CC region). Meanwhile NfeD possesses a sterol-sensing domain (SSD), linked to an OB-fold (OBL), a common ligand-binding domain.
Marta Ukleja, Department of Microbiology CNB-CSIC, said, “We are unraveling the structural puzzle of Flotillin (FloA)—a small membrane protein of the SPFH family—and its partner NfeD, a protein with unique membrane-spanning helices and a cytoplasmic barrel-like domain (Ob-fold). In the lab, we successfully established protocols to heterologously express S. aureus FloA and NfeD in E. coli using a pETDuet vector with His- and FLAG tags.”
A combination of approaches, including fluorescence microscopy, lipid flotation assays, and SEC analysis, demonstrated that the PHB is necessary to recognise and facilitate binding of FloA to STX in the FMM. Alterations to the α-helices in subdomain-1 interrupt this binding, and FloA becomes homogenous throughout the membrane rather than building up at the FMM. FloA and NfeD build up at the FMM due to lipid recognition, with size exclusion chromatography (SEC) then demonstrating that the PHB domain (in conjunction with its CC) is necessary for oligomerisation of FloA.
Ukleja: “Our journey began with negative-staining electron microscopy, giving us a preliminary glimpse of the FloA-NfeD complex. Building on this, we optimised grid preparation for cryo-EM analysis and collected a dataset on the TALOS Arctica 200kV microscope, achieving a 3D reconstruction at 8Å resolution. However, at this resolution, the secondary structure elements of the complex remained elusive. To push the boundaries, we collected high-resolution data using the state-of-the-art Krios Titan 300kV microscope equipped with a Falcon 4 camera. Now, our challenge lies in processing this vast dataset: our lab lacks the computational power and specialised expertise for single-particle 3D reconstruction. To overcome this, we sought support of the Instruct Image Processing Centre (I2PC, part of Instruct-ES).”
The structures of FloA and NfeD were then analysed by Cryo-EM at I2PC. Despite the small size of the proteins, 3D volumes were obtained for FloA (8Å), FloA-NfeD (7.9Å), and 2x-Floa-NfeD (8.8Å) – high resolution structures will be the subject of future studies (a key challenge in cryo-EM is resolving structures below 100 kDa, such as in the case of FloA and NfeD). The CC region of FloA interacts with the OBL domain in NfeD, bending the CC region to create a flexible hook, or tentacle. Purification of dimeric FloA bound to NfeD by SEC allowed the assessment of the dimeric state of FloA and NfeD by cryoEM, yielding a low-resolution map validated by the AF2 model. FloA PHB regions bind to form the FloA dimer (Figure 1), and the CC hooks protruding further away, creating a symmetrical arrangement, and the FloA dimer therefore adopts a “clamp” form.
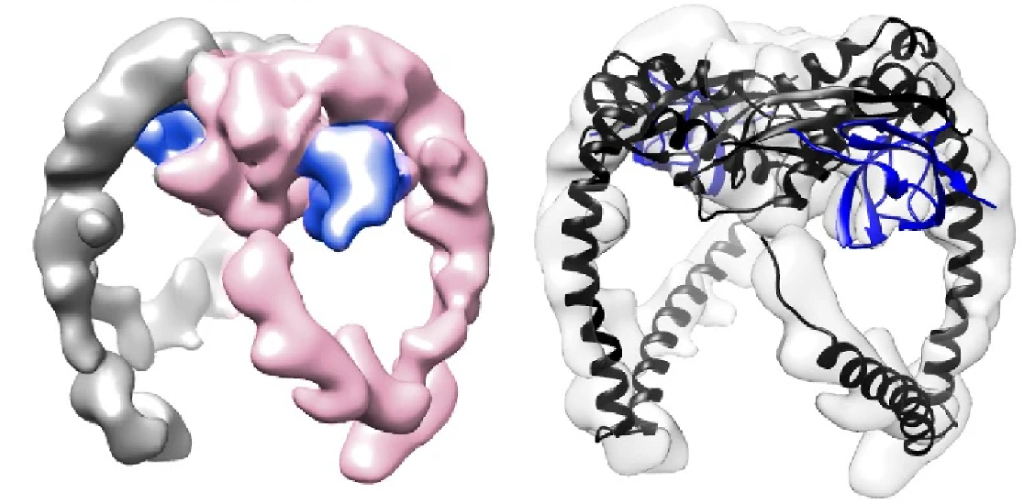
Figure 1. Cryo-EM map of the FloA-NfeD trimer (left). One FloA monomer is labeled in pink and the other FloA monomer in gray. The OBL of NfeD is shown in blue. Cryo-EM map with AF2 overlay (right).
The action of this clamp is the crux of the study, and demonstrates the importance of flotillin for bacterial survival, particularly for S. aureus. Protein structures with CC-OB tentacles are known to prevent the aggregation of denatured proteins, binding to and stabilising unfolded proteins and, upon stabilisation, refolding the client protein to its native state. Gradient fractionation was used to detect that stressed WT cells consistently accumulated insoluble proteins in the FMM, and furthermore, WT bacteria, compared to those with mutated, or deleted FloA, recovered protein solubility over time. Interestingly, recruitment of denatured proteins to the FMM was shown to be purely due to hydrophobic interactions with the FMM resident lipid STX. FloA therefore accumulates at the FMM, alongside insoluble proteins, where it functions to stabilise them for refolding.
To test whether FloA then facilitates the refolding of these proteins, the client protein PBP2a was purified and made to unfold at increasingly higher temperatures. The team found that the addition of FloA or FloA-NfeD (not NfeD alone) led to a revival of protein solubility at these temperatures. They identified that FloA simply stabilised the protein, which then would refold itself to its native state once stabilised, rather than any additional FloA-mediated folding action. Subsequent SEC and cryo-EM analysis showed that the hook region in FloA-NfeD was central to the clamping and stabilisation of PBP2a (Figure 2). This was validated in vivo as PBP2a integrity was compromised when incubated with FloA mutated to remove its CC (hook) domain, compared to WT.
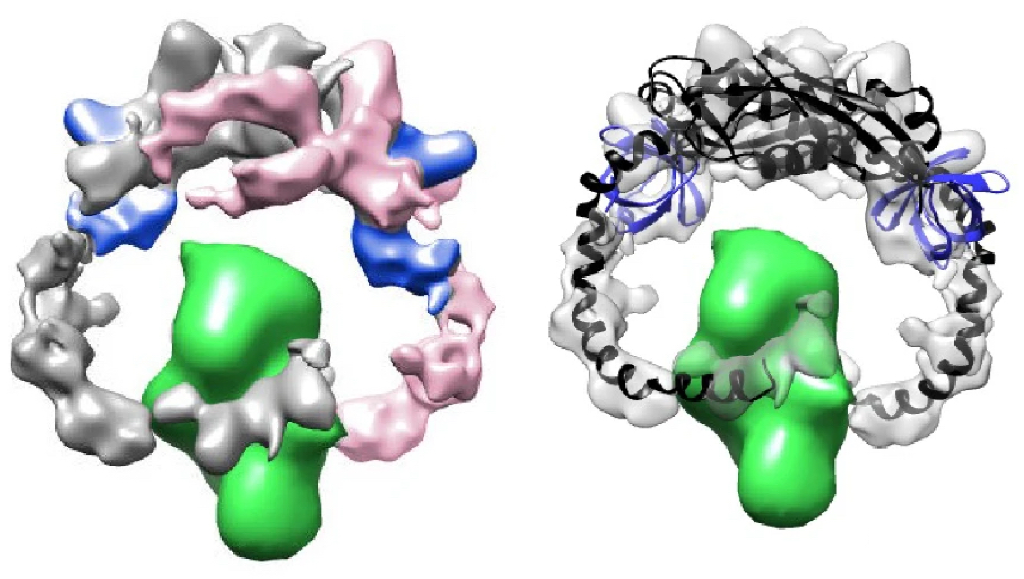
Figure 2. Cryo-EM map of the FloA-NfeD dimer bound to unfolded PBP2a (left). Flexible fitting of the AF2 Multimer prediction is shown in the bottom panels (right). FloA monomer is labeled in pink and the other FloA monomer in gray. The OBL of NfeD is labeled in blue. PBP2a is shown in green.
Ukleja: “The support from Instruct-ERIC I2PC has been instrumental in advancing our project. Their expertise and infrastructure enabled us to process the high-resolution cryo-EM data collected on the FloA-NfeD complex. By collaborating with specialists in single-particle 3D reconstruction, we were able to overcome computational limitations and achieve a structural model, offering new insights into the architecture and function of these critical membrane proteins.”
By facilitating the folding of these client proteins, FloA helps bacteria to withstand stressful environments. For diseases such as MRSA caused by antibiotic resistant S. aureus, this is a field of significant study. Potential treatments could include cholesterol lowering drugs, some of which inhibit the mevalonate pathway (MEV), critical for FMM formation such as Zaragozic acid (ZA), which was shown here to impair FloA oligomerisation in MRSA isolates. In mice, the team found that bacterial load could be reduced 12-fold by ZA treatment , and more than 100-fold in those treated with ZA and standard antibiotic oxacillin, the latter usually finding resistance from MRSA.
This indicates that the FMM, and particularly flotillin, can be a crucial target for drugs in the battle against antibiotic-resistant bacteria. Further structural studies on the precise region of FloA that can best be targeted, and the specific drugs which can be used to inhibit them, will reveal even more about the best mechanisms to beat MRSA and similar bacteria.