07-Jan-2020
Influenza (commonly known as the flu) is an acute, respiratory infection responsible for over 3 million cases of severe illness each year. Most clinical cases of flu are caused by Influenza A viruses, which are also responsible for widespread outbreaks of deadly diseases, such as avian (bird) flu and swine flu. Whilst flu epidemics are a near-annual occurrence, virus mutations that lead to deadly pandemics are unpredictable. To stay one step ahead of flu outbreaks, scientists are trying to decipher the structure and mechanisms of Influenza A viruses, offering scope for the development of new vaccines and antivirals.
Viral-RNA-dependent RNA polymerase (FluPolA) is critical machinery for RNA-containing viruses such as influenza A, as it catalyses the transcription and replication of the viral genome. In a compelling example of integrative structural biology, a team of scientists from the University of Oxford and the Instruct Centre in Belgium have used complementary techniques (including nanobody technology from Instruct-BE) to gain new insights into the replication mechanisms of FluPolA.
FluPolA comprises a complex of three viral proteins: PA, PB1 and PB2, known together as a heterotrimer. Using X-ray crystallography, Fan et al were able to obtain the first, high-resolution structures of FluPolA of human and avian influenza viruses, revealing the dimerisation of the viral protein heterotrimers. To determine whether the dimeric FluPolA structure was important to enzyme activity, mutations were introduced to destabilise the dimer interface. The viral transcription and replication by FluPolA mutants were measured using a mini-replicon assay, showing that the dimeric structure of FluPolA was key in initiating RNA synthesis and stabilised the replication complex.
To investigate the dynamic structure and interactions of FluPolA in its native environment, the team used cryo-electron microscopy (cryoEM) to study the protein binding to an RNA template. CryoEM confirmed that dimer interface of FluPolA was retained in solution and revealed that the 5’-end of the RNA template was bound to FluPolA in a hook conformation in an RNA binding pocket, whilst the 3’-end was more mobile.
To gain further insight into the importance of FluPolA dimerisation, the enzyme was complexed with a nanobody (Nb8205) that reduces dimerisation by binding to a site near to the dimer interface. By comparing the structures of monomeric and dimeric FluPolA-nanobody complexes it was found that dimerisation alters the positions of the proteins in the heterotrimer, aligning the cRNA template for viral-RNA synthesis. The evidence that viral RNA replication is dependent on the dimerisation of FluPolA suggests a target by which virus reproduction might be inhibited. In the future, the characterisation of binding sites that prevent FluPolA dimerisation could lead to the development of antiviral drugs against the flu.
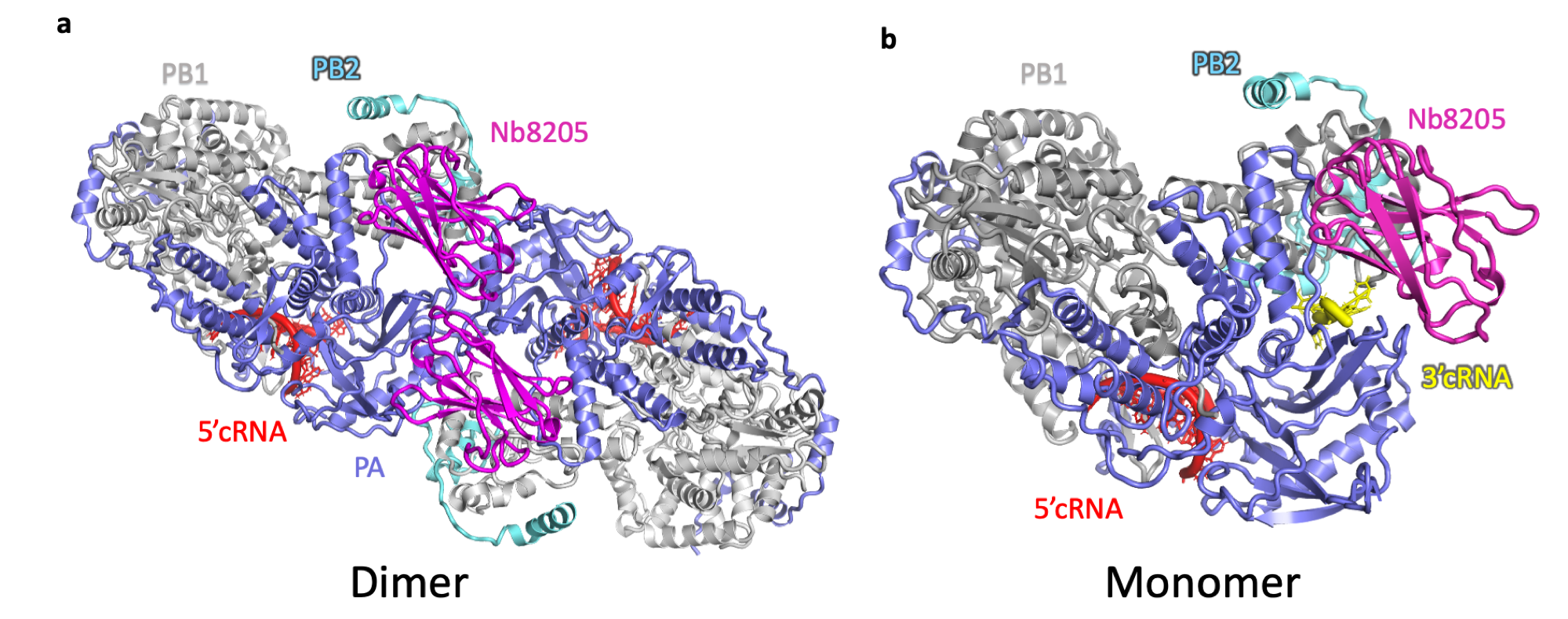
Structures of (a) dimeric and (b) monomeric cRNA-bound human H3N2 FluPolA heterotrimer in complex with Nb8205 solved
by single-particle cryo-EM analysis. In the monomeric structure 3’cRNA (coloured yellow) binds in a narrow groove, that in the dimeric
form of FluPolA is opened up and is not able to accommodate 3’cRNA. The structures in panels a and b are in the same orientation but are
not drawn to scale.
Find out more about the nanobody technology and cryo-EM facilities that are available through Instruct-ERIC.