17-Jan-2025
ß-glucosidase enzymes catalyse the final step of cellulose decomposition, whereby cellobiose, the product of previous enzymatic reactions in the cellulose breakdown process, is converted to glucose (Figure 1A). Whilst ß-glucosidases are attractive biocatalysts due to their wide range of substrates beyond cellobiose, their industrial application is limited by their generally poor thermostability. Thermophilic bacteria, which evolve in extreme environments and possess a suite of highly thermostable enzymes critical to their survival in these environments, are often utilised as a source for more thermostable enzymes for industrial applications. Structures of several ß-glucosidase enzymes originating from various thermophilic bacteria have been deposited in the Protein Databank Europe, each exhibiting a common TIM-barrel fold.
Bgl1, a ß-glucosidase from the thermophilic bacterium Caldicellulosiruptor saccharolyticus, has a favourable half-life and activity at 60°C. It also possesses an intriguing insertion of eight amino acids not present in other structurally characterised ß-glucosidases, rendering it worthy of further study here.
Scientists at Instruct-EL first produced Bgl1 in BL21 DE3 cells, before purifying the enzyme using affinity (His tag, later cleaved with thrombin) and size exclusion chromatography. The activity of the nascent protein was then confirmed by measuring the breakdown of the Bgl1 substrate p-nitrophenyl-β-D-glucopyranoside (pNP-G), by spectroscopic methods (pNP-G absorbs at 410nm). Bgl1 was then concentrated to 16.7 mg/mL for use in automated crystallisation trials using the sitting drop vapour diffusion method. Cocrystals with lactose were also produced to enable the study of the apo and bound forms of the enzyme. These crystals were then studied at the PETRA III synchrotron-radiation source at Instruct-EMBL Hamburg, yielding 1.45Å and 1.95Å resolution structures of the apo and the complex structure respectively (Figure 1B). However, a ß;-D-glucose molecule was bound at the active site of the enzyme instead of lactose.
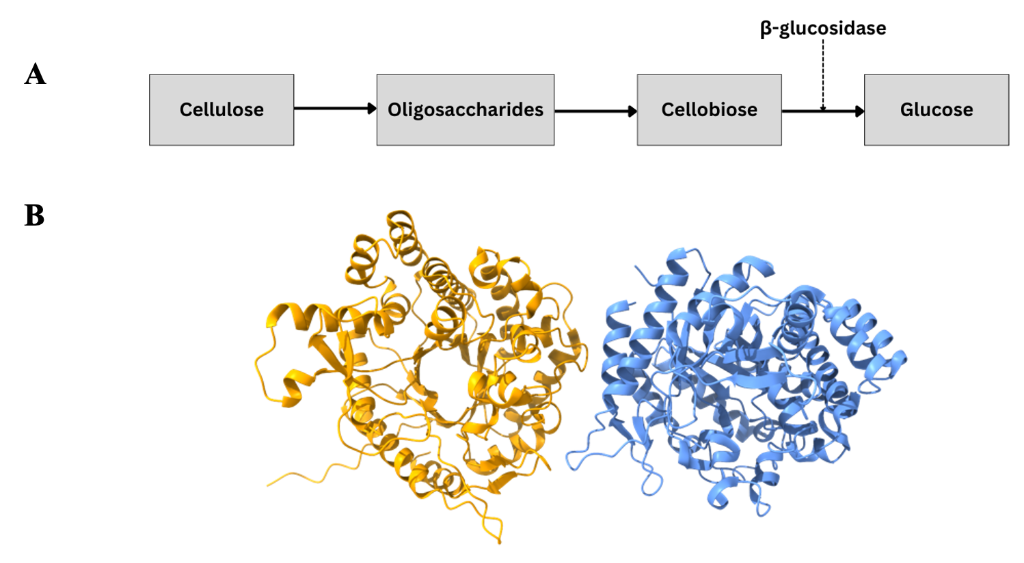
Figure 1: Bgl1 structure and role in cellulose decomposition. A: Schematic of cellulose breakdown process. B: Crystal structure of Bgl1. Identical repeating units shown in yellow and blue.
The structures of Bgl1 and glucose bound Bgl1 exhibit the typical TIM-barrel fold common to the glycosyl hydrolase type 1 (GH1) superfamily to which Bgl1 belongs. Moreover, the key active site residues, Glu163 and Glu361, are the same as other members of the GH1 superfamily. The overall structure of the ß-D-glucose bound and apo forms of Bgl1 are largely the same, with the only differences in structure seen at the active site, where binding of a ß;-D-glucose molecule causes a ~30° rotation in the side chain of Glu163.
The ß-glucosidase with the highest level of sequence and structural homology to Bgl1 belongs to the organism C. cellulovorans. Analysis of the structures of these two proteins reveals that whilst the active site residues are identical, the residues surrounding the active site differ somewhat, resulting in local structural changes depending on the size/charge of the residue. Furthermore, sequence alignments with other Bgl1 homologues reveal two insertions that affect the structure of the enzyme, one of which is unique to Bgl1. Using the tool Deepsite (a tool used for detecting druggable binding pockets, built on deep convoluted neural networks), it was possible to identify a distinct cavity in proximity to the Bgl1 active site made possible due to these insertions. As yet, it is unclear whether this pocket has a functional role, but it is possible that it assists with the binding of the substrate in close proximity to the active site so as to enhance the catalytic efficiency of the enzyme.
Overall, the study demonstrates that for the most part Bgl1 exhibits a structure that is similar to other members of the GH1 superfamily with the exception of two insertions, one of which is unique to Bgl1. Future work will look to use protein engineering approaches to uncover the functional role of this region, if any can be ascribed to it, with a view to furthering our understanding of the enzyme towards industrial biocatalytic applications.