03-Feb-2023
NK cells are part of the immune response – they are lymphocytes equipped with a wide range of activating and inhibitory surface receptors, allowing them to recognise and kill malignant or infected cells.
NKR-P1 is a receptor on NK cells, specifically a CTLR (C-type lectin-like receptor). The receptor particularly binds to LLT1, its endogenous ligand. LLT1 is expressed on several cell types but is especially up-regulated on a number of cancer cells.
The research teams from the Faculty of Science, Charles University, and the Institute of Biotechnology of the Czech Academy of Sciences, BIOCEV research centre in the Czech Republic presented the crystal structure of the NKR-P1 and LLT1 complex. The project was conducted as part of the Instruct-ERIC R&D Pilot Call, which provides funding for pilot studies to deliver preliminary results. Jan Dohnálek, co-author and member of Instruct-CZ, said, “the access to the structural biology infrastructure provided by Instruct-ERIC was fundamental for achieving an important part of the results.”
The paper from Bláha et al. (2022) details how NKR-P1 has a relatively weak affinity to LLT1 but overcomes this with cross-linkages in the binding process.
Initially, two NKR-P1 structures were solved, both glycosylated and deglycosylated, the latter with N-glycans cleaved off using endoglycosidase F1. In the C-type lectin-like domain (CTLD), the structure comprises two α-helices and two antiparallel β-sheets. It was also found that homodimers of NKR-P1 form differently from its homologues, forming via the α1-helix, unlike most other CTLDs. Moreover, deglycosylated NKR-P1 more readily formed homodimers. The crystallisation was performed at STRUBI, part of Instruct-UK.
The crystal structure of the complex of deglycosylated NKR-P1 and LLT1 was solved. The asymmetric unit of the structure contained dimeric LLT1 and two dimeric NKR-P1, with an extra dimer of NKR-P1 bound to the latter.
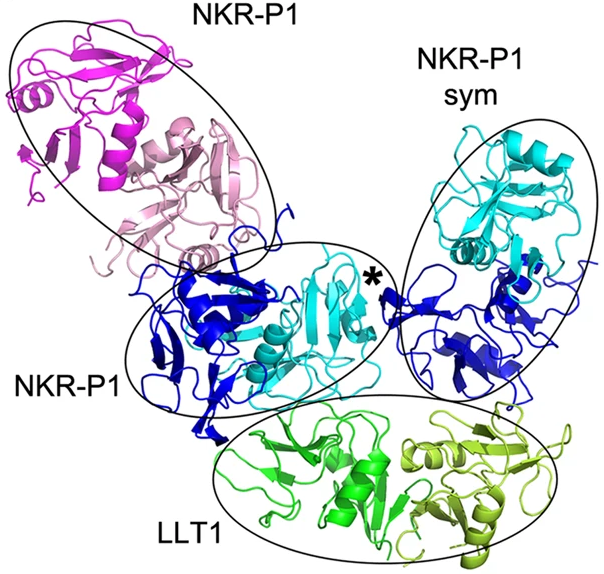
Figure 1. The complex of NKR-P1 and LLT1. Primary interaction is indicated by cyan-NKR-P1 to LLT1; the secondary interaction interface is between blue-NKR-P1 and LLT1. Extra NKR-P1 dimer bound to primary NKR-P1 is shown in pink.
The structure exhibits two different interaction modes. The primary interaction between LLT1 chain B and NKR-P1 chain D is very similar to interactions between NKR-P1 homologues and their ligands. However, the secondary interaction differs significantly from the homologues of the complex.
The main driver of the differences between the interactions is the LLT1 ligand – the primary interface is mostly formed by β3 and β4-sheets of the protein, whereas the secondary binding is conducted through the β2-sheet and α2 helix. In NKR-P1, there is minimal difference in the interaction interfaces for both binding modes.
Through SEC-SAXS conducted at Diamond Light Source (Instruct-UK), the study found that both binding modes were crucial to the formation of higher-order structures of the complex, with particular emphasis on the secondary binding mode in solution and in-cell. It was found that clusters of NKR-P1 form, bound in continuous cross-linked chains with LLT1 (Figure 2), and that the secondary binding mode is indispensable for cross-linking as well as for triggering inhibitory signaling of NKR-P1 in live NK cells. This was in part determined using dSTORM analysis at IGBMC in Strasbourg, Instruct-FR1.
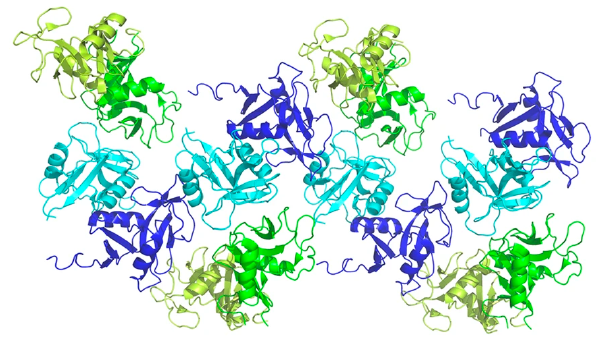
Figure 2. Four adjacent asymmetric units within the NKR-P1:LLT1 complex crystal. The NKR-P1 (blue and cyan) and LLT1 (green and lemon) dimers alternate in primary (cyan and green) and secondary (blue and lemon) interactions, forming a chain-like structure.
This discovery of a novel multimerisation of the NK cell receptor:ligand complex was a key part of the study, explaining how the ligand binding can overcome the relatively low affinity. This discovery could significantly impact how immune recognition is carried out by NK cells in humans.
Ondřej Vaněk, the corresponding author, said, “Instruct was really instrumental to [the project’s] success, both the provided access to various platforms and the R&D Pilot Project funding helping us to achieve the aim of publishing more than just a structure itself, but also its biological implications for immune recognition.”