13-Jun-2023
Transposases are crucial enzymes in catalysing the process of transposition of mobile DNA elements called transposons across loci in genomes. This reorganisation of DNA caused by the displacement of DNA mobile elements across genomes is central to long term evolutionary and diversification processes, but also leads to modifications in gene expression. It was for example shown to be involved in some cancer development but also to stimulate antibiotic resistance genes in bacteria, and as a result is linked to several infectious diseases in humans.
The most abundant type of transposons are insertion sequences (ISs). A widespread class of IS is the IS21 family, which has been found in drug-resistant strains of E. coli and S. aureus, and led to the evolution of the Yersinia pathogens.
The structure of IS21 is similar to many transposons – two gene sequences which both code for their respective transposases (IstA and IstB in the case of the IS21 family), flanked by terminal inverted repeats (TIRs). In this study, Spínola-Amilibia et al (2023) use biochemical and structural approaches to define the molecular determinants by which IstA catalyses efficient DNA transposition.
First, a biochemical assay showed that IstA is not sufficient to complete the transfer of IS21 from a donor DNA strand to a supercoiled target strand. IstB is also needed for the integration reaction. Additionally, IstA requires specific TIR repeats (two repeats on both the left and right) in order to successfully transpose IS21 – random sequences are insufficient.
IstA, like many transposases, forms a large complex to bridge the transposon ends for insertion. Once IstA engages with the TIR repeats of IS21, it assembles into a stable higher-order oligomer with 2-fold symmetry. It was also found that IstA can bind both transposon ends simultaneously, leading to enhanced efficiency in formation of the oligomeric complex.
Cryo-EM was then used to identify the self-assembled three-dimensional tetramer formed by IstA. The structure of the tetramer can be seen in Figure 1a. This cryo-EM experiment took place at Diamond Light Source, a facility in the Instruct-UK centre, and initial data processing was performed at I2PC (Instruct-ES centre), following a successful application to get access to advanced structural biology services through Instruct-ERIC.
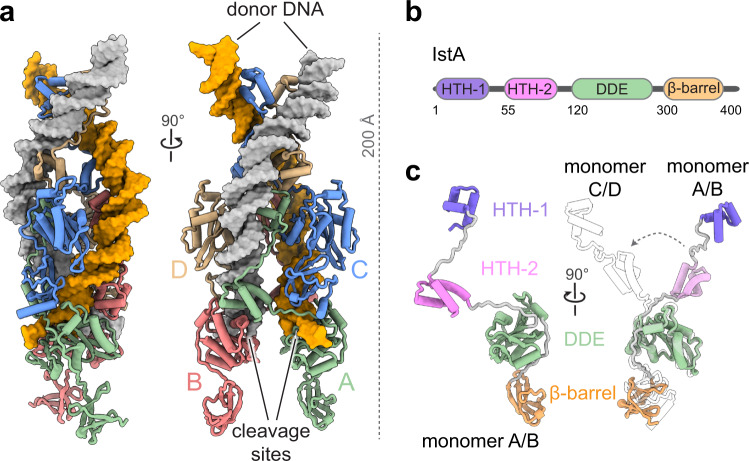
Figure 1. A) Cryo-EM structure of IstA tetramer bound to donor DNA. B) Organisation of IstA. C) Structure of pair of IstA monomers
The cryo-EM structure revealed that the IstA monomer is composed of four domains. Figures 1b and 1c show these: two N-terminal helix-turn-helix (HTH) regions, the catalytic DDE domain, which carries out the transposition process. Finally, a small ß-barrel and c-terminus completes the structure.
Within this complex, the upper IstA monomers (C and D) dimerise directly through the HTH regions, whereas the lower monomers nearer the TIR cleavage sites are joined by the DNA binding site of one monomer to the DDE module of the other, and vice versa. Therefore, the catalytic components (DDE) of the lower dimers have much more of a functional role in the complex, binding to the transposon in trans. The often-reported role of this bridge is to prevent DNA breakages as the transposon ends are bound.
As mentioned previously, the IstA requires two repeats in the TIR when it binds. The lower catalytic dimer binds to the outer repeat, the higher structural dimer binding to the inner repeat of the TIR, specifically the right side (R1 outer, R2 inner). These points are specifically recognised by IstA, and it is at this point that the donor DNA is bent sharply, causing it to wrap around the tetramer for insertion then into the target strand.
The central role that transposases play cancer and drug-resistance make them a key target for health research worldwide. This study gives a crucial indication into how they operate with regard to the abundant IS21 transposons, and will give rise to greater understanding and study of these enzymes in the future.