15-Dec-2021
As the COVID-19 pandemic remains, the conversation around variants continues with it. The emergence of new variants brings the risk that vaccines developed against earlier strains of SARS-CoV-2 might not be efficient against new strains. Understanding the difference between variants of SARS-CoV-2 at a structural level provides a foundation for drug discovery and vaccine development.
Early in 2021, researchers at the Wellcome Centre for Human Genetics utilised technology at Diamond Light Source, part of Instruct-UK. They studied the effectiveness of vaccines and natural sera (taken from patients infected by an early strain of SARS-CoV-2) against the variants Alpha and Beta, compared to the early Victoria strain of the virus initially discovered in Wuhan. They found that neutralisation of the Beta variant was lower compared to Alpha.
A follow-up study has been published, again utilising the advanced technology available at Diamond Light source, to understand the antigenic distance between Beta and other SARS-CoV-2 variants, namely: Alpha, Gamma and Delta, as well as the Victoria strain.
Liu et al (2021), identified 27 monoclonal antibodies (mAbs) from blood serum of those who had tested positive for the Beta variant of COVID-19. These 27 showed potent neutralising activity from a panel of 674 - selected based on their binding to the Receptor Binding Domain (RBD) of the Beta Spike (S) protein and achieving >90% neutralisation against Beta.
These mAbs were then tested against the following viruses (with indicated changes to the RBD) in a live virus neutralisation: Alpha (N501Y), Beta (K417N, E484K, N501Y), Gamma (K417T, E484K, N501Y), Delta (L452R, T478).
Although the majority of these neutralizing mAbs retain their neutralisation potential against Alpha and Gamma variants, many of them significantly reduce or lose their effect against the Victoria and the Delta strains. In comparison to their neutralising efficiency against Beta, mAbs experienced total knock out of neutralising potential against each variant in the following proportions: Victoria (10/27), Alpha (5/27), Gamma (1/27), Delta (15/27).
Of the three Beta mutations, the most commonly targeted residue change by the mAbs was N501Y (11/27), followed by E484K (6/27), and K417N (3/27). Variants that share one or more of these residue changes were more potently neutralised by Beta serum.
As the Gamma variant carries similar mutations to Beta (sharing E484K and N501Y), the neutralisation efficiency is closer than that of the other strains. At the other end of the scale, the Delta variant has a high antigenic distance from Beta (not sharing any residue mutations), demonstrated by more than 55% of mAbs being knocked out in the neutralisation titre.
The study utilised X-Ray diffraction techniques at Diamond Light Source, analysing crystallised structures of the Spike RBD in complex with each of the mAbs. This provided the range of structures available to the researchers, giving a visual overview of how each mAb interacts with beta RBD and interfere with the binding to ACE2, as well as demonstrating where the mutations occur for each variant."
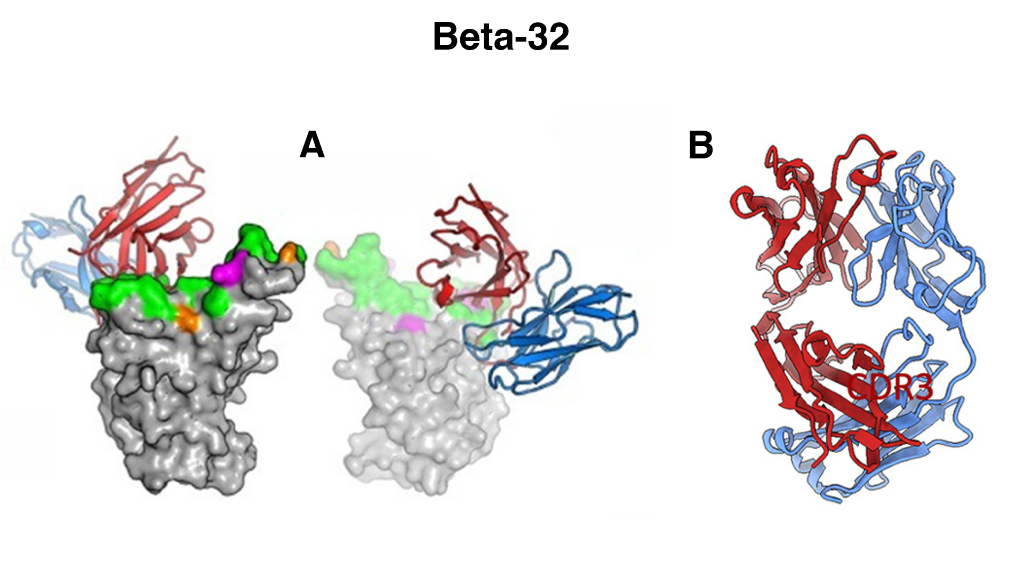
Figure 1. Figure 4. Overall structures of Beta-32 (A) Front and back views of Beta RBD/Beta Fab complexes. Fabs drawn as ribbons with HC red and LC blue, and RBDs as grey surfaces with ACE2 footprint in green, mutation sites of the Beta variant in magenta and Delta variant in orange. Crystal structure of Beta-32 Fab (B) with HC red and LC blue.
One interesting example neutralising mAb highlighted in this study is Beta-32. Beta-32 binding to Beta S was investigated both by crystallisation and by Cryo-EM. Surprisingly, Beta-32 was found to be highly potent against all variants – despite its binding via the 501 residue that is mutated in Alpha, Beta and Gamma variants, as it interacts via a different angle to other mAbs. This is a potential target, therefore, for drug development due to its unique approach to RBD binding. The structure of Beta-32 is shown in Figure 1. Fig1 (A) is Beta 32 Fab (antigen binding fragment) binding to beta RBD of S protein and (B) is the beta-32 Fab alone.
By understanding the location of mutation on SARS-CoV-2 variants and binding for the most potent neutralising mAbs using structural biology techniques, further research has a platform on which to design potential combatants against the virus.