25-Aug-2022
Actin is one of the most highly conserved proteins in eukaryotes, playing a pivotal role in cell migration, endocytosis, and muscle contraction. It is the coordinated polymerisation and disassembly of actin filaments that allow it to play such a fundamental role in cells. However, different organisms have quite different methods and machineries for facilitating actin filament dynamics.
Kotila et al. (2022) studied actin filaments of Leishmania major, a trypanosomatid parasite responsible for severe disease in humans, and compared it to mammalian actin. The aim of the study was to understand the differences in actin depolymerisation between both groups, and the enzymes and cofactors involved – which would in turn demonstrate the evolution of actin and its regulatory machinery.
The initial part of the study was to assess L. major actin (LmActin) through microfluidics, generating long actin filaments from LmActin subunits in solution. The LmActin was able to form long filaments when polymerising at the barbed end, even when combined with rabbit skeletal muscle α-actin (RbActin), albeit at a much slower rate. The team also found that LmActin depolymerised 5x more rapidly than mammalian actin.
The next step of the study was using Cryo-EM at the University of Helsinki, part of Instruct-Finland. Cryo-EM was used in order to understand the molecular principles behind the differences in actin structure and conformation – generating structural images and obtaining advanced structural data. The resulting images showed that the structure of parasitical and mammalian actin was very well conserved, however a number of deletions and alterations in the structure may explain the different in depolymerisation rates. This is shown particularly in the core of LmActin, and also in the pointed end, between the subunits of actin – weaker connections may explain the more rapid depolymerisation rates.
In order to examine the role of actin-regulatory machinery on the actin filament dynamics, filaments of LmActin and RbActin were saturated with cofilin (Figure 1), a key actin regulator that promotes actin filament diassembly. Cofilin is known to be inefficient at disassembly in yeast and mammals, so often requires a cofactor (Aip1 the most common). However, Aip1 is not found in L. major. The resulting experiment showed that as expected, mammalian Cofilin bound to RbActin but did not rapidly depolymerise in absence of cofactors such as Aip1, however LmCofilin disassembled LmActin 100x more efficiently.
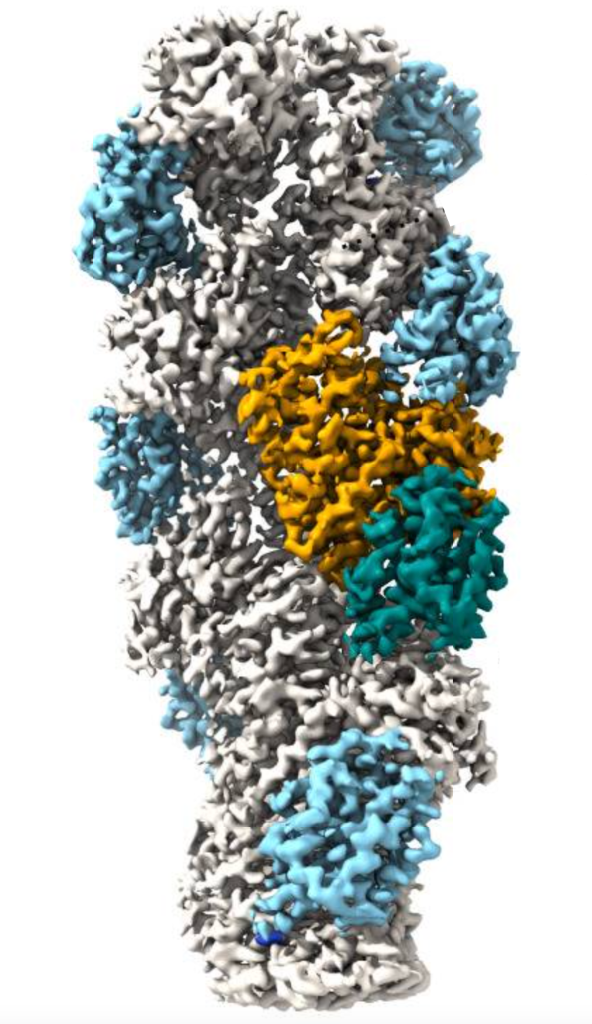
Figure 1. LmActin filament bound to by LmCofilin (orange / blue-green section)
Further Cryo-EM examination found that key differences in the N and C-termini of cofilins and actin filament structures between both groups were responsible for this significant difference.
The study provides a foundation for future systematic mutagenesis and molecular dynamic simulation studies, as well as open new avenues for developing specific inhibitors against parasitic actins, particularly in devastating trypanosomes such as L. major.
Read the full text of the article here: Kotila et al., Structural basis of rapid actin dynamics in the evolutionarily divergent Leishmania parasite. Nature Communications (2022). 10.1038/s41467-022-31068-y