22-Sep-2022
In cells, DNA is wound around complexes made of eight histone proteins, forming nucleosomes. These nucleosomes make up the standard configuration of DNA, chromatin.
The assembly of nucleosomes requires the specific binding of 8 histones: an (H3-H4)2 tetramer, as well as two H2A-H2B dimers. Nucleosome assembly was until now always described as a sequential process in which H3-H4 were first deposited on the DNA followed by H2A-H2B deposition. However, landmark results from Corbeski et al. (2022) show that the DNA repair factor APLF is able to assemble and deposit the histone octamer directly on the DNA.
The incumbent understanding of this binding was that several chaperone proteins were central to the process, binding the histones to form the octamer complex, which then wrapped the DNA strand around to form the nucleosome. However, landmark results from Corbeski et al (2022) show that the action of a single chaperone protein can in fact guide the entire process.
APLF (aprataxin and polynucleotide kinase–like factor) is known to be a DNA repair factor, providing the scaffold for other proteins to join broken DNA ends.
What the team from Utrecht University uncovered, however, was that the C-terminus acidic domain of APLF (APLFAD) had individual binding sites for both H3-H4 and H2A-H2B, binding all complexes simultaneously. This separated it from other histone chaperones, which typically bind either H2A-H2B or H3-H4. The use of Nuclear Magnetic Resonance techniques at the Utrecht NMR Group at Utrecht University and small-angle scattering at EMBL Hamburg were crucial to uncover this novel binding mode.
Hugo van Ingen, lab leader, said, “Thanks to Instruct-ERIC / Instruct-ULTRA we could push the structural characterization of the APLF-histone complex beyond the crystallized APLF fragment and perform integrative NMR and small-angle scattering studies, via access to the bioSAXS facility at DESY/EMBL Hamburg. This provided the key structural information to understand APLF’s function.”
The crystal structure of a minimal complex was solved at the Netherlands Cancer Institute (NKI) in collaboration with the Sixma group. The crystal showed that through the two binding sites, APLFAD is found to be unique among chaperone proteins in that it can bind both H3-H4 and H2A-H2B, and in their native nucleosome configuration.
They found that aromatic anchor residues at the APLFAD binding sites for H3-H4 and H2A-H2B were crucial for histone octamer formation, by mutating the aromatic anchor residues present at these sites. Upon mutation of these residues, it was shown by that binding by APLFAD to histone octamers was reduced by five-fold and increased non-native histone-histone contacts, highlighting how crucial these aromatic residues are on the binding and chaperoning of the histone octamers. It also indicated that APLFAD more readily bound to histone octamers, than just H3-H4 and H2A-H2B individually.
Using the NMR, SAXS and cross-linking mass-spectrometry results, they found that the full APLFAD binds the entire histone octamer, except for the central dyad region (Figure 1). This is the area which then binds more readily to DNA during nucleosome assembly.
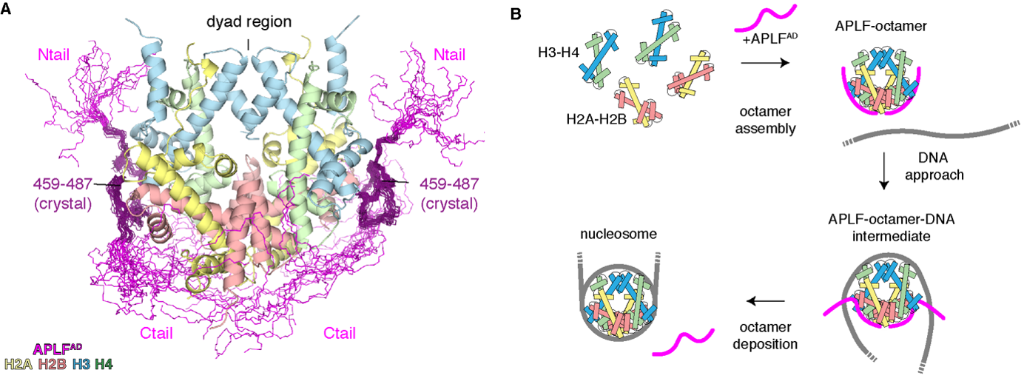
Figure 1. (A) Integrative structure of the histone octamer complex bound by APLFAD based on crystallography, NMR, SAXS and XL-MS data. Both the H3-H4 and H2A-H2B regions are bound by the C-tail region of APLFAD, covering the entire octamer but for the central dyad region. This region more effectively binds to DNA, ensuring that APLFAD effectively deposits the histones for nucleosome assembly (B).
Ivan Corbeski, first author, then established that APLFAD operates as a histone chaperone by depositing octamers onto DNA to form nucleosomes. Using nucleosome assembly and quantitation assays in collaboration with the Mattiroli group at the Hubrecht Institute, it was shown that the addition of APLFAD to the histone octamers and DNA mix significantly increased nucleosome formation.
To summarise, APLFAD uniquely binds both the histone complexes H3-H4 and H2A-H2B simultaneously to form the histone octamer. It covers the full octamer except for the region which binds to DNA, allowing nucleosome formation when the octamer is deposited on the DNA. This all challenges the previous paradigm that nucleosome formation was a multi-step process involving multiple chaperones – instead APLFAD indicates that this process can happen in one step, which is a process that might be shared by other histone chaperones.
This crucial paper demonstrates for the first time that histone octamer formation and nucleosome assembly can be uncoupled processes. It also suggests that APLF can capture histone octamers and re-deposit these complexes on the DNA following repair, which would allow for maintenance of epigenetic information. The importance of histone formation and DNA repair means that this discovery could have significant benefits both for immune responses and cancer biology in the future.