07-Jun-2023
The glucocorticoid receptor (GR) is a ubiquitously expressed nuclear hormone receptor. It is activated through endocrine signalling by cortisol, and is of fundamental importance for development, skeletal growth, and other bodily functions. The structure of GR includes a large, disordered amino-terminal domain (NTD), a DNA-binding domain (DBD) and a ligand-binding domain (LBD).
The structure of GR has been studied in intense detail over the years, particularly to determine how it binds to both ligands and DNA, and the differences it carries from other endocrine receptors (ER).
This study from Postel et al (2022) aimed to determine the structure of GR, and explore how the various domains differ and communicate with one another. This was done with the experience of the team at IGBMC (Instruct-FR1), and with AstraZeneca, showcasing a fantastic collaborative effort between academia and industry!
The study presents the crystal structures of GR (residues 385–777, encompassing the DBD and LDB) in complex with the agonists velsecorat and fluticasone furoate, a natural GBS and a co-regulator peptide. These are the first high-resolution multidomain structures of a steroid receptor, and they reveal how the receptor forms a unique architecture on the DNA.
The protein constructs, shown in Figure 1, were of the full GR protein (a), the construct missing part of the NTD (b, stabilised by several mutants), and the construct missing both the NTD and DBD (c).
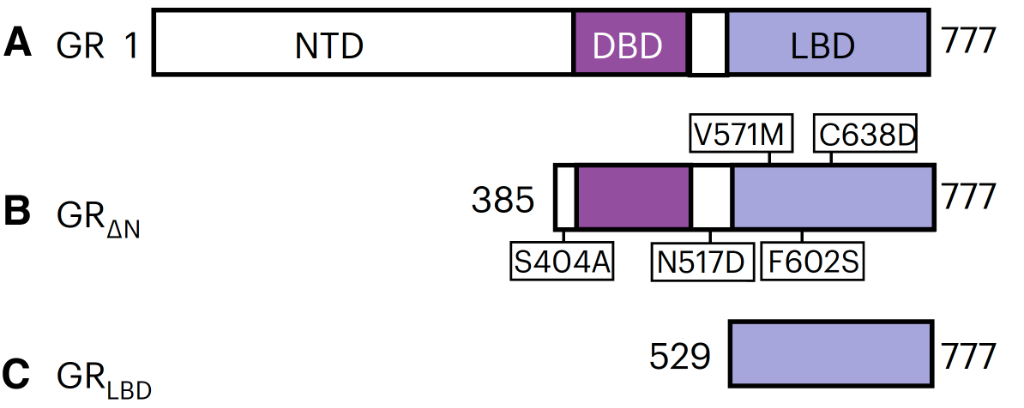
Figure 1. The full GR protein (a), the construct missing part of the NTD (b, stabilised by several mutants), and the construct missing both the NTD and DBD (c).
The purified receptors were then incubated with double-stranded DNA (dsDNA) and co-regulator peptide to obtain quaternary complexes. The highest resolution quaternary structure was GRΔN in complex with: velsecorat (-vel), a GR-binding sequence from serum and glucocorticoid-regulated kinase-1 (SGK1), and a peptide (residues 134–154) from the co-regulator peroxisome proliferator-activated receptor γ co-activator 1-α (PGC1α). This whole complex is referred to as GRΔN-vel-SGK1- PGC1α.
The complex forms a tetrahedral shape, with the LBD and DBD at opposite corners, as the LBD dimer is bound asymmetrically in the centre of the DBD dimer. This is illustrated in Figure 2. In the crystal lattice, there are two potential LBD dimers: either head to tail or head to head (mediated by H1 in this instance).

Figure 2. The LBD dimer bound in the centre of the DBD dimer, which is then bound to the DNA strand.
The study found that the quaternary structure domain arrangement is different from all other quaternary nuclear receptor structures reported to date
The structures were also examined in complex with several different ligands (velsecorat, fluticasone furoate or dexamethasone). This demonstrated structural changes between binding of different ligands (velsecorat protected the H1 region), but also demonstrated that hydrogen-deuterium exchange carried out at different rates depending on the ligand.
This provides valuable and unique insight into the structure of GR in complex with DNA and ligands, and also demonstrates the mechanism by which domains communicate with one another in this state. This will give a significant boost to drug development around GR, and demonstrates how academic and industrial partners can collaborate to identify long-term health solutions.