18-Dec-2023
Helicases are ubiquitous molecular motor proteins involved in all aspects of DNA metabolism. Their primary function is to catalyse duplex DNA unwinding in an ATP-dependent fashion. A subset of DNA helicases is also known to unwind non-canonical secondary DNA structures, called G-quadruplexes (G4s), which induce genome instability if left unresolved.
RECQ4 protein is a helicase implicated in G4 metabolism, which contains an intrinsically disordered region (IDR) at the N-terminus of its sequence. Owing to their conformational flexibility and dynamics, IDRs are thought to expand the biomolecular functionality of ordered proteins and domains.
The research teams of Konstantinos Tripsianes from CEITEC, Masaryk University, and Lumír Krejčí from Faculty of Science, Masaryk University, has uncovered a positively charged polyelectrolyte in the IDR of RECQ4 helicase. The physicochemical properties of the IDR account for the conformational plasticity in diverse functions and coacervation with oppositely charged polyelectrolytes such as G4 structures. Their findings offer interesting perspectives in understanding RECQ4 function in the cell linked to the partitioning of proteins into condensates.
The use of solution NMR spectroscopy at Instruct-CZ, combined with biochemical, biophysical, and optical methods, was instrumental in deciphering how the IDR is able to switch between phases, conformational states and binding partners. The integrated approach allowed to bridge microscopic measurements and macroscopic observations into a three-step kinetic model of liquid-liquid phase separation, perhaps one of the first attempts to describe this molecular phenomenon that it is implicated in many cellular functions.
In their paper, published in Nature Communications, the authors found that the IDR resembles a Swiss army knife offering RECQ4 the molecular tools to participate in different cellular processes.
The structural, kinetic, and thermodynamic profile of its interactions revealed distinct modes of binding depending on the partner. On one hand, the IDR remains highly flexible in polyelectrolyte interactions with nucleic acids. The disordered state in these complexes minimizes the entropic penalty of binding, resulting in relatively strong but non-specific interactions. On the other hand, the IDR forms a stable helix when it interacts with replication protein A (RPA), the major protein that binds to single-stranded DNA in eukaryotic cells. The induced folding comes at an entropic cost, resulting in an interaction characterized by high specificity but low affinity. NMR competition experiments show that the two binding modes are mutually exclusive.
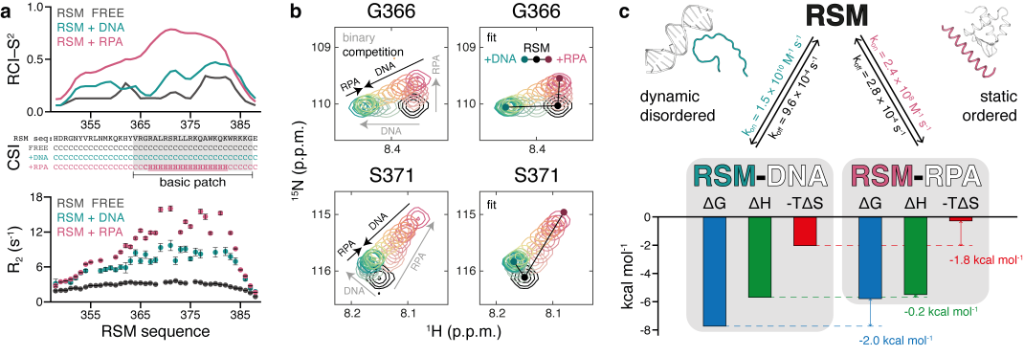
Figure 1. Distinct and competitive binding modes of RECQ4 IDR (RSM) to DNA and RPA
(a) Order parameters and secondary structure derived from backbone chemical shifts and 15N R2 relaxation rates measured for free RSM (black) and for RSM in complex with DNA (cyan) or RPA (pink). (b) Competitive binding of DNA and RPA to RSM, observed by 2D NMR titrations. Gray arrows indicate peak trajectories upon binding of each partner and black arrows indicate peak trajectories upon competition with the other partner (left). Fitted spectra following 2D line shape analysis of the competition binding (right). (c) Summary of kinetic, thermodynamic, and structural parameters of RSM interactions.
Interestingly, when the RECQ4 IDR binds to G-quadruplexes, liquid-liquid phase separation (LLPS) occurs, similar to when droplets of oil appear in a mixture of oil and water. By integrating equilibrium and kinetic binding data in a global numerical model, a three-step model of charge-driven coacervation is deduced. The oppositely charged IDR and G4 molecules form a complex in the solution that follows a rapid nucleation-growth mechanism leading to a dynamic equilibrium between dilute and condensed phases. Additional experiments using differential labeling of the interacting partners confirmed the exchange of RECQ4 IDR and G4 between the solution and droplets.
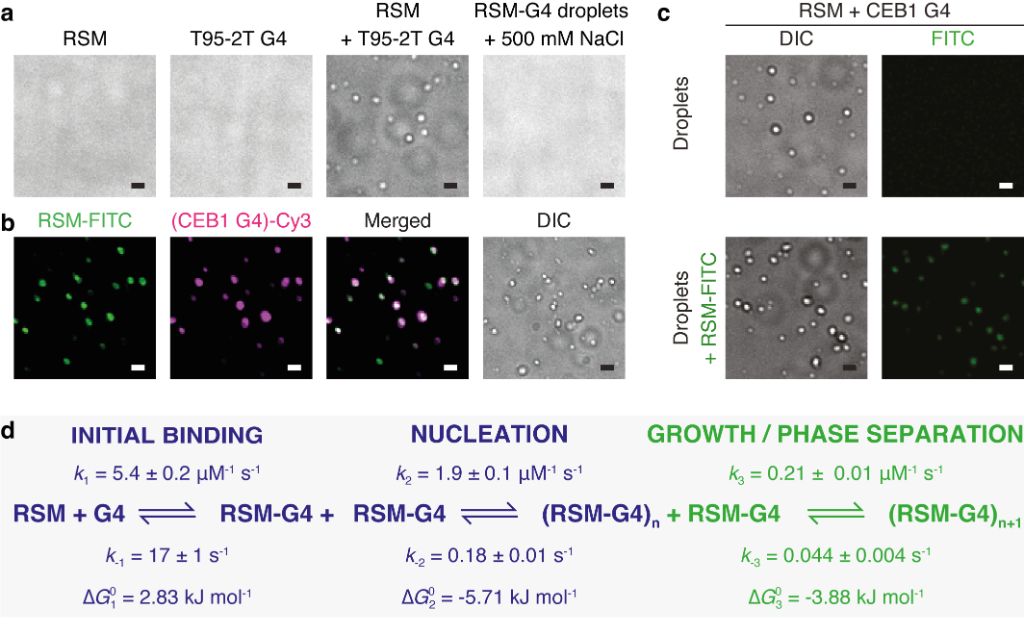
Figure 2. Coacervation between RECQ4 IDR (RSM) and G-quadruplexes
(a) RSM (10 μM), parallel T95-2T G4 (10 μM) or their mixture were analysed for droplet formation using DIC microscopy. The addition of 500 mM NaCl dissolves the droplets. (b) Droplet formation and colocalisation of 5 mol % FITC-labeled RSM (10 μM) and 5 mol % Cy3-labeled CEB1 G4 (10 μM). (c) Droplet formation (upper panels) and colocalisation (lower panels) of 5 mol % FITC-labeled RSM with preformed droplets of RSM and CEB1 G4 (10 μM each). In all images scale bar = 1 μm. (d) The minimal kinetic model describing the sRSM-G4 coacervation includes three main steps: (i) initial binding, (ii) formation of stable nuclei and (iii) growth leading to phase separation. The rate constants for the individual steps were derived from combining equilibrium titration data with transient stopped-flow kinetics in a global numerical model.
The dynamic nature of coacervation and the ability to explore the two extremes of the structural continuum of complexes in selecting partners (RPA or DNA) suggest a regulatory role for the IDR to enable diverse RECQ4 functions in DNA replication and DNA repair in general.